About RaptorX
RaptorX is a protein structure prediction server developed by Xu group, excelling at predicting 3D structures for protein sequences without close homologs in the Protein Data Bank (PDB). Given an input sequence, RaptorX predicts its secondary and tertiary structures, contacts, solvent accessibility, disordered regions and binding sites. RaptorX also assigns some confidence scores to indicate the quality of a predicted 3D model: P-value for the relative global quality, GDT (global distance test) and uGDT (un-normalized GDT) for the absolute global quality, and modeling error at each residue.
RaptorX performance in CASP9
RaptorX excels at the alignment of hard targets, which have less than 30% sequence identity with solved structures in PDB. As shown in the below figure, blindly tested on the 50 hardest CASP9 template-based modeling targets, RaptorX outperforms all the CASP9 participating servers including those using consensus and refinement methods.
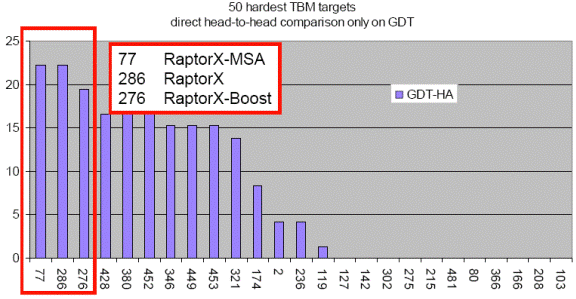
The above figure shows the performance of top 25 groups on the 50 hardest CASP9 TBM targets. X: group numbers. Y: the number of top 25 groups outperformed by a given group. The figure is taken from the CASP9 assessor’s presentation at http://predictioncenter.org/casp9/doc/presentations/CASP9_TBM.pdf
RaptorX performance in CASP10
The below figure shows the top 10 groups (out of ~200) on the most difficult CASP10 template-based modeling targets. RaptorX is the only server among the top 10. The other 9 are human groups, which can make use of all server results.
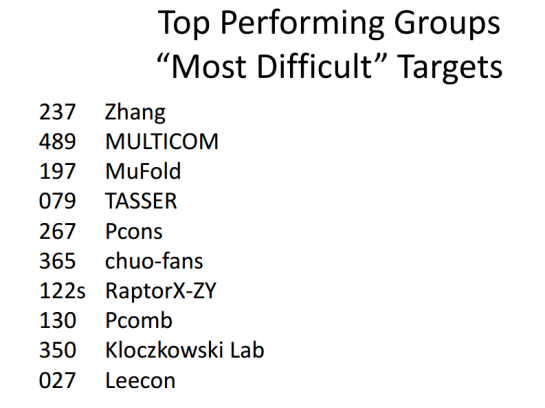
The left column is the group number with 's' indicating a server group and the right column is the group name.
Citing RaptorX
Server Paper
Morten Källberg, Haipeng Wang, Sheng Wang, Jian Peng, Zhiyong Wang, Hui Lu & Jinbo Xu. Template-based protein structure modeling using the RaptorX web server. Nature Protocols 7, 1511–1522, 2012.Method Papers for tertiary structure prediction
Ma J, Wang S, Zhao F, Xu J. Protein threading using context-specific alignment potential. Bioinformatics (Proceedings of ISMB 2013). 2013 Jul 1;29(13):i257-65. doi: 10.1093/bioinformatics/btt210.Jianzhu Ma, Jian Peng, Sheng Wang and Jinbo Xu. A conditional neural fields model for protein threading. Bioinformatics (Proceedings of ISMB 2012), 2012.
Jian Peng and Jinbo Xu. RaptorX: exploiting structure information for protein alignment by statistical inference. PROTEINS, 2011.
Jian Peng and Jinbo Xu. A multiple-template approach to protein threading. PROTEINS, 2011.
Jian Peng and Jinbo Xu. Low-homology protein threading. Bioinformatics (Proceedings of ISMB 2010 , 2010.
Jian Peng and Jinbo Xu. Boosting protein threading accuracy. In the Proceedings of the 13th International Conference on Research in Computational Molecular Biology (RECOMB), Lecture Notes in Computer Science. Vol. 5541, pp. 31-45, 2009. Springer.
Method Paper for secondary structure prediction
Wang Z, Zhao F, Peng J, Xu J. Protein 8-class secondary structure prediction using conditional neural fields. Proteomics. 2011 Oct;11(19):3786-92. doi: 10.1002/pmic.201100196. Epub 2011 Aug 31.Method Paper for contact prediction
Zhiyong Wang and Jinbo Xu. Predicting protein contact map using evolutionary and physical constraints by integer programming. Bioinformatics (Proceedings of ISMB 2013), 2013. Also see an Extended versionJianzhu Ma, Sheng Wang, Zhiyong Wang and Jinbo Xu. Protein Contact Prediction by Integrating Joint Evolutionary Coupling Analysis and Supervised Learning. RECOMB 2015, Lecture Notes in Computer Science ), Volume 9029, 2015, pp 218-221.
Related Papers
Feng Zhao, Jian Peng and Jinbo Xu. Fragment-free approach to protein folding using conditional neural fields. Bioinformatics (Proceedings of ISMB 2010), 2010.Feng Zhao, Jian Peng, Joe DeBartolo, Karl F. Freed, Tobin R. Sosnick and Jinbo Xu. A probabilistic and continuous model of protein conformational space for template-free modeling. Journal of Computational Biology, 2010.
RaptorX Storage for Publication
In general jobs will be deleted from the server 6 months after completion due to limited storage capacity. We do, however, recognize that user may want to refer to their results from the server in published work and further, make these results available to the public.
To request that a specific set of results be stored permanently on the server and/or be made publicly available please contact us.