1.Introduction
The documentation describes how to submit jobs to the RaptorX Contact server, as well as retrieve and interpret results. For detailed explanation of the algorithms underlying the server please see the relevant papers listed at http://raptorx2.uchicago.edu/ContactMap/about/.
2.Job Submission and User Account
- Job submission
-
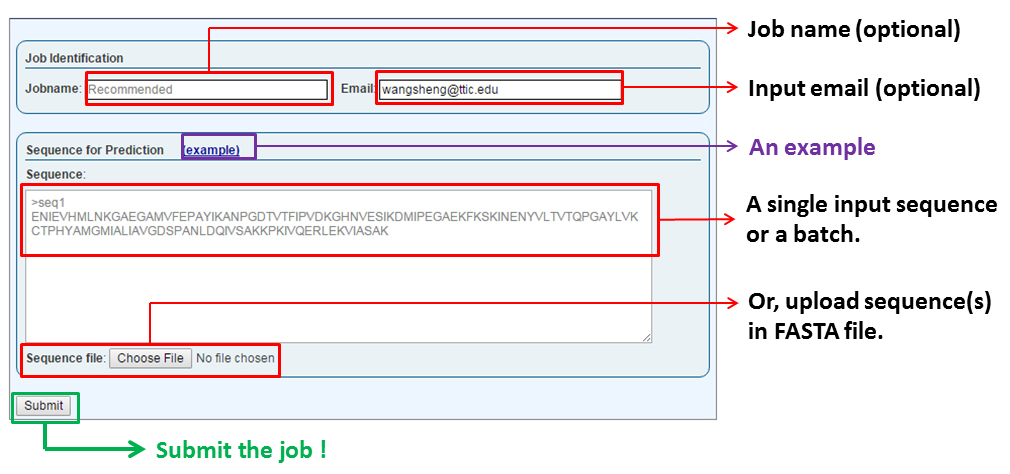
The "Job Identification" section allows users to provide a job name (default is 'my job') and an email address to be used for notification when the job is done (if you are already logged in this field will be pre-filled). An email is NOT required, but STRONGLY RECOMMEND since it can be used to retrieve the results of all your jobs. The email provided in submission also serves as the username of a user.
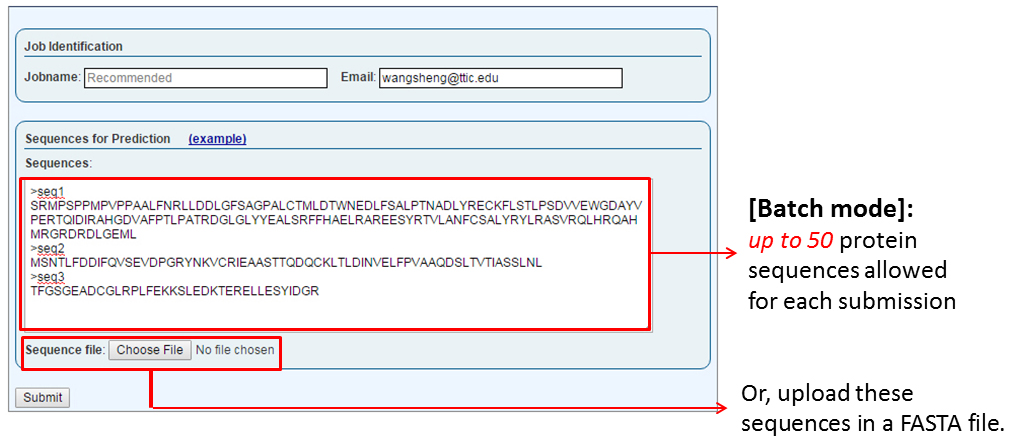
The "Sequences for Prediction" secction is where a user submits one or more sequences (the batch mode) in the FASTA format. The sequence(s) can either be supplied by copy-and-pasting into the text box or by uploading a FASTA-formatted text file. Successful submission will redirect the user to a page showing the status of this submitted job.

Jobs can also be submitted using a command-line program curl according to the above format. Note that for curl submission, only one sequence is allowed at a time, and only terms with underline are required. The job URL is returned on screen after successful submission.
- User account and job retrieval
-
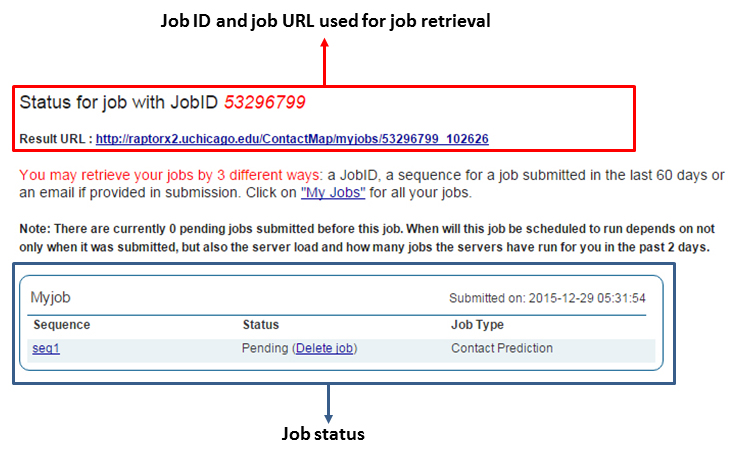
We strongly suggest that you provide an email address in submission, which will facilitate the retrieval of your jobs. A new user account identified by your email address is automatically created when you submit your first job. In the case that you do not provide an email address, you may check the status of your job by its server-assigned JobID or its sequence. In case that you cannot find your jobs by all these methods, please contact the RaptorX team.
When your account is first created (after you submitted your first job) you are automatically logged into the server on the machine you are using at that point. If your login session has expired or you want to access your account from a different machine, you may go to the server front page http://raptorx2.uchicago.edu and supply your account email in the login field on the right. Few minutes after submission of the form you will receive an email notification containing a hyperlink to the page containing the jobs for the account.
- Notes
-
A single job may contain at most 50 sequences and each user can have no more than 500 sequences in pending at any time. Further, the results of a job are guaranteed to be stored for only 14 days after the job is completed, although empirically all the current jobs are stored for two years. To store your jobs for a much longer time for publications, please contact the RaptorX team through the "Inquiry & Bug Report" link.
3.Job Monitoring and Job Availability

The "Job Identification" section allows users to provide a job name (default is 'my job') and an email address to be used for notification when the job is done (if you are already logged in this field will be pre-filled). An email is NOT required, but STRONGLY RECOMMEND since it can be used to retrieve the results of all your jobs. The email provided in submission also serves as the username of a user.

The "Sequences for Prediction" secction is where a user submits one or more sequences (the batch mode) in the FASTA format. The sequence(s) can either be supplied by copy-and-pasting into the text box or by uploading a FASTA-formatted text file. Successful submission will redirect the user to a page showing the status of this submitted job.

Jobs can also be submitted using a command-line program curl according to the above format. Note that for curl submission, only one sequence is allowed at a time, and only terms with underline are required. The job URL is returned on screen after successful submission.
We strongly suggest that you provide an email address in submission, which will facilitate the retrieval of your jobs. A new user account identified by your email address is automatically created when you submit your first job. In the case that you do not provide an email address, you may check the status of your job by its server-assigned JobID or its sequence. In case that you cannot find your jobs by all these methods, please contact the RaptorX team.

When your account is first created (after you submitted your first job) you are automatically logged into the server on the machine you are using at that point. If your login session has expired or you want to access your account from a different machine, you may go to the server front page http://raptorx2.uchicago.edu and supply your account email in the login field on the right. Few minutes after submission of the form you will receive an email notification containing a hyperlink to the page containing the jobs for the account.
A single job may contain at most 50 sequences and each user can have no more than 500 sequences in pending at any time. Further, the results of a job are guaranteed to be stored for only 14 days after the job is completed, although empirically all the current jobs are stored for two years. To store your jobs for a much longer time for publications, please contact the RaptorX team through the "Inquiry & Bug Report" link.
There are three ways to retrieve your jobs: (a) by JobID, (b) by email address, and (c) by sequence.
- By email address
-
The user needs to be logged in to the server. Refer to Section 2 of this manual for login instructions. Once logged in to the server, selecting "My Jobs" in the menu at the top of the page display a job overview page similar to the one depicted below. Here the status of each prediction in the job is given along with overall information of the predictions being done for each sequence submitted. To track the job status in real-time simply refresh the page and the completion status of the prediction submitted for each sequence in a job will be updated. Clicking on a sequence name will take the user to the result page for this sequence.
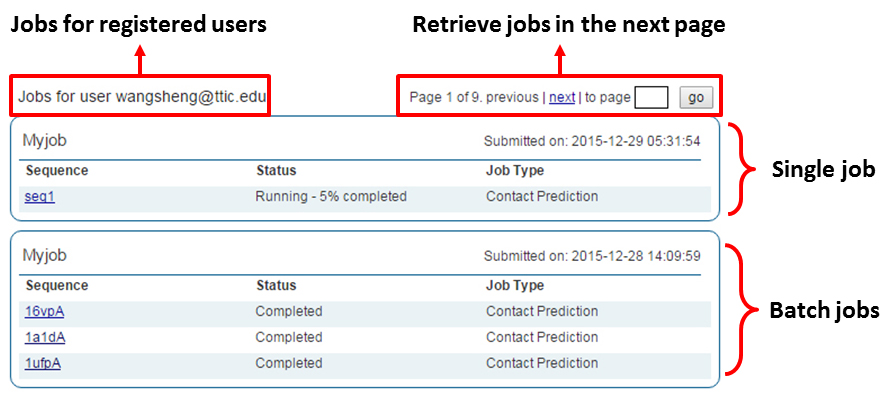
- By job ID or sequence
-
Use the "Retrieve Results by JobID" or the "Job status" link. The remaining procedure is similar to job retrieval by email address.
4.Interpretation of Results
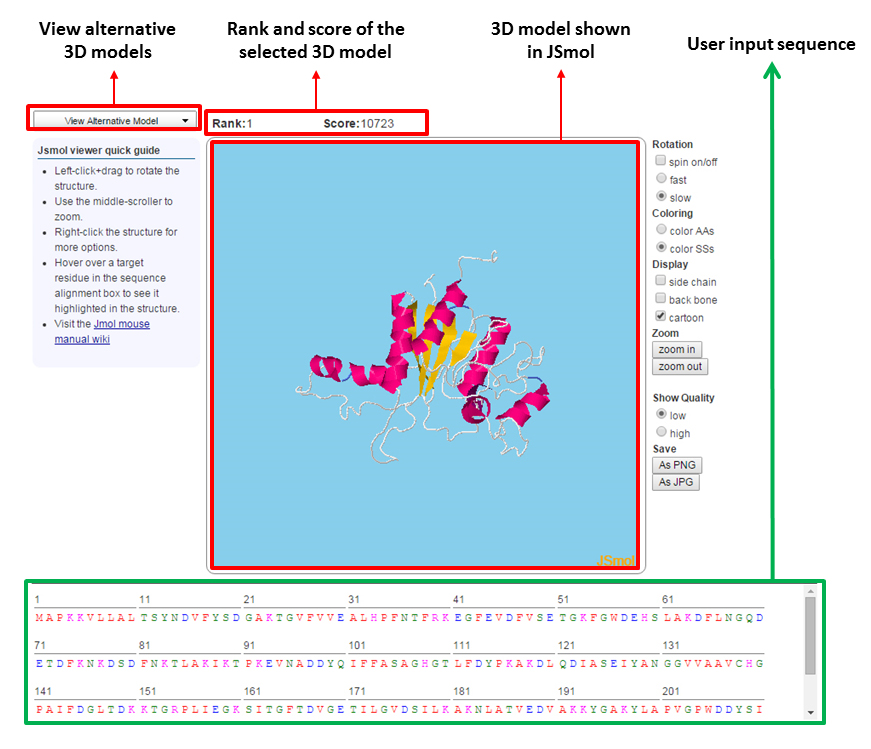
We provide three prediction results: (a) a predicted contact map, (b) a contact result file, and (c) five predicted 3D models assisted by the predicted contacts.
- Predicted contact map
-
For the contact map, we display the probability of two residues being in contact (i.e., their C-beta distance falling in the range [0,8A]). Higher probabilities are represented by darker color.
- Contact result file
-
The contact prediction results in a '.RR' formatted file for each input protein sequence. This file follows the format used in CASP ROLL, click this link for details.
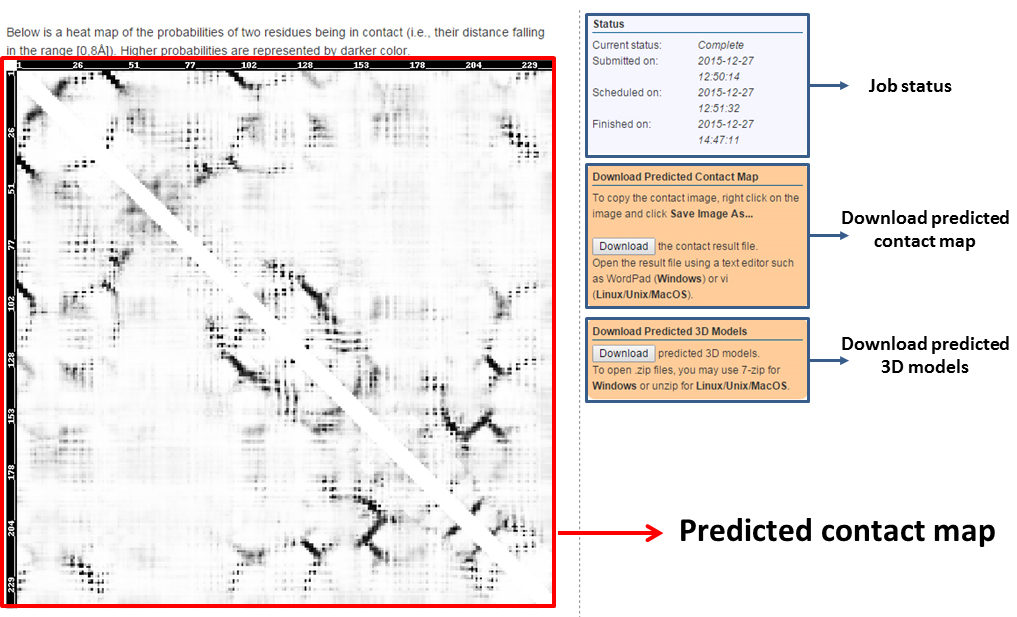
- Predicted 3D models
-
Five predicted 3D models are generated with the assistance of the predicted contacts by CNS_solve program. We also provide the energy score for each predicted 3D model.